Selection plus FunPDBe Study Case of Q13426
pip install --upgrade pdb-profiling
Import Packages & Settings
from pdb_profiling import default_config
default_config("C:/GitWorks/pdb-profiling/test/demo")
from pdb_profiling.processors import SIFTS, PDB, Base
from pdb_profiling.utils import DisplayPDB
from tqdm.notebook import tqdm
from pandas import concat, DataFrame
# 链层面筛选过滤 (Chain Level Filtering)
# 设置SISTS.chain_filter条件: UNK_COUNT < SEQRES_COUNT,下面展示默认值(Default value is shown below)
SIFTS.chain_filter = '''
UNK_COUNT < SEQRES_COUNT
and ca_p_only == False
and identity >=0.9
and repeated == False
and reversed == False
and OBS_COUNT > 20'''
valid filters:
| Column Name | Type | Explaination |
|---|---|---|
| identity | float | provided by SIFTS: sequence identity of PDB Entity SEQRES with UniProt Isoform (0-1) |
| is_canonical | bool | whether the UniProt Isoform is the canonical isoform defined by UniProt-KB |
| sifts_range_tag | str | Safe or Insertion or Deletion or InDel (example) |
| reversed | bool | whether there is reversed mapped range in the aspect of UniProt Isoform Sequence (example) |
| repeated | bool | whether there is repeated mapped range in the aspect of UniProt Isoform Sequence (example) |
| InDel_sum | int | SEQRES residues that fall into the range of Insertion or Deletion or InDel of the PDB Chain Instance |
| unp_len | int | the length of the UniProt Isoform Sequence |
| BINDING_LIGAND_COUNT | int | the residues that binding to ligands(including carbohydrate polymer) of the PDB Chain Instance |
| OBS_COUNT | int | the observed/modelled (with coordinates) residues of the PDB Chain Instance |
| OBS_RATIO_SUM | float | the sum of the observed/modelled (with coordinates) residues’s ratio of the PDB Chain Instance |
| NON_COUNT | int | the count non-standard residues of the PDB Entity (including UNK) |
| SEQRES_COUNT | int | the count of the residues in SEQRES |
| STD_COUNT | int | the count of the standard residues of the PDB Entity |
| UNK_COUNT | int | the count of the UNK residues of the PDB Entity |
| ca_p_only | bool | whether the PDB Entity only contains C-alpha atom for each residue |
| OBS_STD_COUNT | int | the count of the observed standard residues of the PDB Chain Instance |
# PDB条目层面筛选过滤(Entry Level Filtering)
# 设置SISTS.entry_filter条件,下面展示默认值(Default value is shown below)
SIFTS.entry_filter = '''
(experimental_method in ["X-ray diffraction", "Electron Microscopy"] and resolution <= 3) or
experimental_method == "Solution NMR"
'''
valid filters:
| Column Name | Type | Explanation |
|---|---|---|
| resolution | float/nan | (pdb-101-explanation) |
| experimental_method_class | str | (pdb-101-explanation) |
| experimental_method | str | x-ray, nmr, em, other |
| multi_method | bool | whether the PDB entry was determined by multiple method |
| revision_date | date | as name said |
| deposition_date | date | as name said |
demo = SIFTS('Q13426')
Select Monomeic Protein
Implement
PDBe RESTful API(PDBe Entry&SIFTS)
%time df1 = demo.pipe_select_mo().result()
df1[df1.select_tag.eq(True)].T
Wall time: 654 ms
| 4 | |
|---|---|
| UniProt | Q13426 |
| chain_id | A |
| entity_id | 1 |
| identity | 0.99 |
| is_canonical | True |
| pdb_id | 3ii6 |
| struct_asym_id | A |
| pdb_range | [[1,203]] |
| unp_range | [[1,203]] |
| Entry | Q13426 |
| range_diff | [0] |
| sifts_range_tag | Safe |
| repeated | False |
| reversed | False |
| InDel_sum | 0 |
| new_pdb_range | [[1,203]] |
| new_unp_range | [[1,203]] |
| conflict_pdb_index | {"60":"A","134":"I"} |
| conflict_pdb_range | [[60,60],[134,134]] |
| conflict_unp_range | [[60,60],[134,134]] |
| unp_len | 336 |
| BINDING_LIGAND_COUNT | 0 |
| BINDING_LIGAND_INDEX | [] |
| OBS_COUNT | 201 |
| OBS_INDEX | [[1, 201]] |
| OBS_RATIO_SUM | 201 |
| ARTIFACT_INDEX | [] |
| NON_COUNT | 0 |
| NON_INDEX | [] |
| SEQRES_COUNT | 203 |
| STD_COUNT | 203 |
| STD_INDEX | [[1, 203]] |
| UNK_COUNT | 0 |
| UNK_INDEX | [] |
| ca_p_only | False |
| molecule_type | polypeptide(L) |
| OBS_STD_INDEX | ((1, 201),) |
| OBS_STD_COUNT | 201 |
| RAW_BS | 0.587555 |
| RAW_BS_IG3 | 0.587555 |
| resolution | 2.4 |
| experimental_method_class | x-ray |
| experimental_method | X-ray diffraction |
| multi_method | False |
| revision_date | 20110713 |
| deposition_date | 20090731 |
| 1/resolution | 0.416667 |
| id_score | -65 |
| select_tag | True |
| select_rank | 1 |
DisplayPDB(dark=True).show('3ii6', range(1,3))
| Asymmetric unit of 3ii6 | Biological assembly 1 of 3ii6 | Biological assembly 2 of 3ii6 |
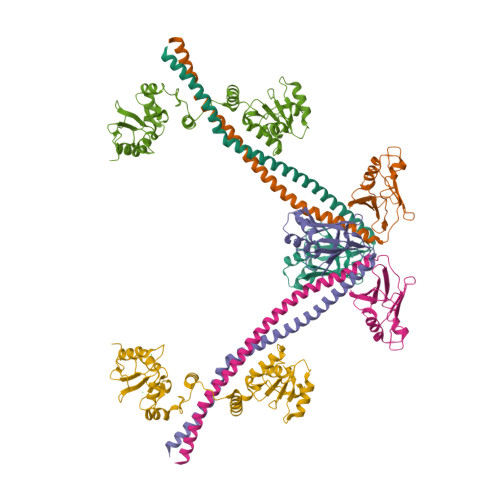 | 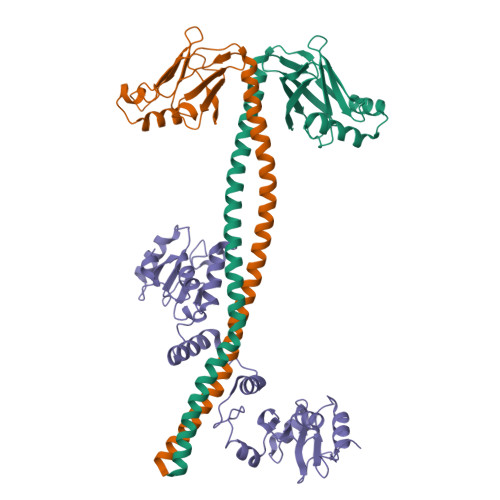 | 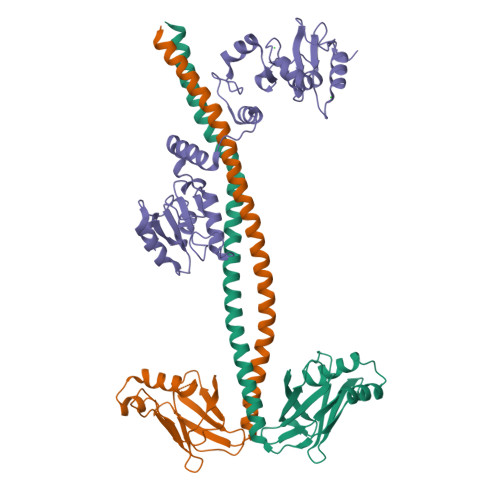 |
Prepare for Residue-Level Mapping
record = df1[df1.select_tag.eq(True)].iloc[0]
mapping_df = PDB(record['pdb_id']).get_expanded_map_res_df(
record['UniProt'],
record['new_unp_range'],
record['new_pdb_range'],
struct_asym_id=record['struct_asym_id']).result()
mapping_df
| unp_residue_number | residue_number | UniProt | author_insertion_code | author_residue_number | chain_id | entity_id | multiple_conformers | observed_ratio | pdb_id | residue_name | struct_asym_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | Q13426 | 1 | A | 1 | NaN | 1 | 3ii6 | MET | A | |
| 1 | 2 | 2 | Q13426 | 2 | A | 1 | NaN | 1 | 3ii6 | GLU | A | |
| 2 | 3 | 3 | Q13426 | 3 | A | 1 | NaN | 1 | 3ii6 | ARG | A | |
| 3 | 4 | 4 | Q13426 | 4 | A | 1 | NaN | 1 | 3ii6 | LYS | A | |
| 4 | 5 | 5 | Q13426 | 5 | A | 1 | NaN | 1 | 3ii6 | ILE | A | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 198 | 199 | 199 | Q13426 | 199 | A | 1 | NaN | 1 | 3ii6 | LEU | A | |
| 199 | 200 | 200 | Q13426 | 200 | A | 1 | NaN | 1 | 3ii6 | ASN | A | |
| 200 | 201 | 201 | Q13426 | 201 | A | 1 | NaN | 1 | 3ii6 | ALA | A | |
| 201 | 202 | 202 | Q13426 | 202 | A | 1 | NaN | 0 | 3ii6 | ALA | A | |
| 202 | 203 | 203 | Q13426 | 203 | A | 1 | NaN | 0 | 3ii6 | GLN | A |
203 rows × 12 columns
Detecting Homomeric Interaction
also annotated by
PISA&Interactome3D
from pdb_profiling.processors.i3d.api import Interactome3D
Interactome3D.pipe_init_interaction_meta().result()
%time df2 = demo.pipe_select_ho(run_as_completed=True, progress_bar=tqdm).result()
df2[df2.i_select_tag.eq(True)]
HBox(children=(FloatProgress(value=0.0, max=5.0), HTML(value='')))
Wall time: 2.47 s
| entity_id_1 | chain_id_1 | struct_asym_id_1 | struct_asym_id_in_assembly_1 | asym_id_rank_1 | model_id_1 | molecule_type_1 | surface_range_1 | interface_range_1 | entity_id_2 | ... | select_rank_2 | in_i3d | unp_range_DSC | best_select_rank_score | second_select_rank_score | unp_interface_range_1 | unp_interface_range_2 | i_group | i_select_tag | i_select_rank | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | A | A | A | 1 | 1 | polypeptide(L) | [[15,35],[37,47],[49,108],[110,126],[128,227]] | [[19,21],[30,31],[33,33],[51,54],[132,134],[13... | 1 | ... | 4 | True | 1.0 | 0.250000 | 0.250000 | ((5, 7), (16, 17), (19, 19), (37, 40), (118, 1... | ((5, 7), (16, 17), (19, 19), (37, 40), (118, 1... | (Q13426, Q13426) | True | 10 |
| 7 | 1 | A | A | A | 1 | 1 | polypeptide(L) | [[1,21],[23,201]] | [[11,16],[89,91],[103,103]] | 1 | ... | 7 | False | 1.0 | 1.000000 | 0.142857 | ((11, 16), (89, 91), (103, 103)) | ((1, 1), (3, 3), (25, 25), (121, 121), (124, 1... | (Q13426, Q13426) | True | 4 |
| 8 | 1 | B | B | B | 1 | 1 | polypeptide(L) | [[1,19],[21,35],[37,76],[82,94],[96,201]] | [[117,118],[121,121],[124,124]] | 1 | ... | 7 | False | 1.0 | 0.200000 | 0.142857 | ((117, 118), (121, 121), (124, 124)) | ((117, 118), (121, 121), (124, 124)) | (Q13426, Q13426) | True | 11 |
| 10 | 1 | A | A | A | 1 | 1 | polypeptide(L) | [[1,21],[23,201]] | [[5,7],[15,17],[19,19],[37,40],[119,121],[123,... | 1 | ... | 5 | True | 1.0 | 1.000000 | 0.200000 | ((5, 7), (15, 17), (19, 19), (37, 40), (119, 1... | ((5, 7), (16, 17), (19, 19), (38, 40), (117, 1... | (Q13426, Q13426) | True | 2 |
| 11 | 1 | A | A | A | 1 | 1 | polypeptide(L) | [[1,21],[23,201]] | [[7,7],[9,9],[14,15],[17,17],[19,19],[80,80]] | 1 | ... | 2 | False | 1.0 | 1.000000 | 0.500000 | ((7, 7), (9, 9), (14, 15), (17, 17), (19, 19),... | ((7, 7), (9, 9), (14, 15), (17, 17), (80, 80)) | (Q13426, Q13426) | True | 1 |
| 16 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[1,35],[37,178]] | [[57,62],[65,65],[98,98],[101,107]] | 1 | ... | 6 | True | 1.0 | 0.166667 | 0.111111 | ((57, 62), (65, 65), (98, 98), (101, 107)) | ((1, 1), (3, 3), (23, 25), (30, 33), (46, 46),... | (Q13426, Q13426) | True | 15 |
| 21 | 1 | B | B | B | 1 | 1 | polypeptide(L) | [[1,17],[19,33],[35,35],[37,41],[43,203]] | [[145,145],[148,149],[152,152],[155,156],[158,... | 1 | ... | 6 | True | 1.0 | 0.166667 | 0.166667 | ((145, 145), (148, 149), (152, 152), (155, 156... | ((145, 145), (148, 149), (152, 152), (155, 156... | (Q13426, Q13426) | True | 12 |
| 22 | 1 | A | A | A | 1 | 1 | polypeptide(L) | [[1,178]] | [[7,7],[9,9],[15,17]] | 1 | ... | 9 | True | 1.0 | 0.111111 | 0.111111 | ((7, 7), (9, 9), (15, 17)) | ((57, 57), (62, 62), (64, 64)) | (Q13426, Q13426) | True | 21 |
| 23 | 1 | A | A | A | 1 | 1 | polypeptide(L) | [[1,178]] | [[166,166],[169,170],[173,174]] | 1 | ... | 9 | False | 1.0 | 0.111111 | 0.111111 | ((166, 166), (169, 170), (173, 174)) | ((166, 166), (169, 170), (173, 174)) | (Q13426, Q13426) | True | 22 |
9 rows × 114 columns
Detecting Heteromeric Interaction
also annotated by
PISA&Interactome3D
%time df3 = demo.pipe_select_he(run_as_completed=True, progress_bar=tqdm).result()
df3[df3.i_select_tag.eq(True)]
HBox(children=(FloatProgress(value=0.0, max=3.0), HTML(value='')))
Wall time: 2.23 s
| entity_id_1 | chain_id_1 | struct_asym_id_1 | struct_asym_id_in_assembly_1 | asym_id_rank_1 | model_id_1 | molecule_type_1 | surface_range_1 | interface_range_1 | entity_id_2 | ... | select_tag_2 | select_rank_2 | in_i3d | best_select_rank_score | second_select_rank_score | unp_interface_range_1 | unp_interface_range_2 | i_group | i_select_tag | i_select_rank | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[15,33],[35,35],[37,47],[49,108],[110,126],[1... | [[169,169],[172,173],[176,177],[179,180],[183,... | 2 | ... | True | 1 | True | 1.0 | 0.25 | ((155, 155), (158, 159), (162, 163), (165, 166... | ((465, 466), (469, 470), (473, 473), (476, 477... | (Q13426, Q0D2I5) | True | 1 |
| 11 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[15,33],[35,35],[37,47],[49,108],[110,126],[1... | [[169,169],[172,173],[176,177],[179,180],[183,... | 2 | ... | True | 1 | True | 1.0 | 0.25 | ((155, 155), (158, 159), (162, 163), (165, 166... | ((105, 106), (109, 110), (113, 113), (116, 117... | (Q13426, Q0D2I5-2) | True | 1 |
| 12 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[15,33],[35,35],[37,47],[49,108],[110,126],[1... | [[169,169],[172,173],[176,177],[179,180],[183,... | 2 | ... | True | 1 | True | 1.0 | 0.25 | ((155, 155), (158, 159), (162, 163), (165, 166... | ((468, 469), (472, 473), (476, 476), (479, 480... | (Q13426, Q0D2I5-4) | True | 1 |
| 13 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[15,33],[35,35],[37,47],[49,108],[110,126],[1... | [[169,169],[172,173],[176,177],[179,180],[183,... | 2 | ... | True | 1 | True | 1.0 | 0.25 | ((155, 155), (158, 159), (162, 163), (165, 166... | ((469, 470), (473, 474), (477, 477), (480, 481... | (Q13426, Q0D2I5-5) | True | 1 |
| 14 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[15,33],[35,35],[37,47],[49,108],[110,126],[1... | [[169,169],[172,173],[176,177],[179,180],[183,... | 2 | ... | True | 1 | True | 1.0 | 0.25 | ((155, 155), (158, 159), (162, 163), (165, 166... | ((106, 107), (110, 111), (114, 114), (117, 118... | (Q13426, Q0D2I5-6) | True | 1 |
| 15 | 1 | A | A | AA | 2 | 2 | polypeptide(L) | [[15,33],[35,35],[37,47],[49,108],[110,126],[1... | [[169,169],[172,173],[176,177],[179,180],[183,... | 2 | ... | True | 1 | True | 1.0 | 0.25 | ((155, 155), (158, 159), (162, 163), (165, 166... | ((468, 469), (472, 473), (476, 476), (479, 480... | (Q13426, Q0D2I5-7) | True | 1 |
| 41 | 1 | C | C | C | 1 | 1 | polypeptide(L) | [[1,21],[23,201]] | [[150,150],[153,154],[157,158],[161,161],[164,... | 2 | ... | True | 1 | True | 1.0 | 0.50 | ((150, 150), (153, 154), (157, 158), (161, 161... | ((763, 771), (774, 775), (778, 778), (800, 800... | (Q13426, P49917) | True | 1 |
7 rows × 117 columns
Collecting Residue-Level Annotation From FunPDBe via PDBe Graph API
PDBe-KB consortium, PDBe-KB: a community-driven resource for structural and functional annotations, Nucleic Acids Research, Volume 48, Issue D1, 08 January 2020, Pages D344–D353, https://doi.org/10.1093/nar/gkz853
| Partner resource (Reference) | Resource leader | Type of annotations | Number of PDB entries |
| COSPI-Depth (21) | M. S. Madhusudhan | Residue depth | 141 097 |
| P2rank (6) | D. Hoksza | Binding site predictions | 138 892 |
| Arpeggio (15) | T. Blundell | Ligand interactions | 117 023 |
| 3DComplex (14) | E. D. Levy | Interaction interfaces | 111 555 |
| DynaMine (19) | W. Vranken | Backbone flexibility predictions | 98 548 |
| POPSCOMP (20) | F. Fraternali | Solvent accessibility | 77 578 |
| AKID (11) | M. Helmer-Citterich | Kinase-target predictor | 41 492 |
| ChannelsDB (9) | R. Svobodova | Molecular channels | 25 351 |
| CATH-FunSites (13) | C. Orengo | Functional site predictions | 23 975 |
| canSAR (7) | B. al-Lazikani | Druggable pocket predictions | 17 804 |
| FoldX (17) | L. Serrano | Energetic consequences of mutations | 3778 |
| ProKinO (10) | N. Kannan | Curated regulatory sites | 3673 |
| 14–3-3-Pred (12) | G. Barton | Binding site predictions | 1941 |
| CaMKinet (in preparation) | M. Kumar | Curated PTM sites | 1076 |
| M-CSA (5) | J. Thornton | Curated catalytic sites | 919 |
| 3DLigandSite (8) | M. Wass | Binding site predictions | 910 |
| Missense3D (18) | M. Sternberg | Mutations in Human Proteome | 0* |
| MetalPDB (16) | A. Rosato | Curated metal binding sites | 0* |
| ELM (24) | T. Gibson | Short linear motifs | 0* |
 |
pdb_ob = PDB(record['pdb_id'])
pdb_ob
<PDB 3ii6>
funpdbe_df = pdb_ob.fetch_from_pdbe_api('graph-api/pdb/funpdbe_annotation/', Base.to_dataframe).result()
funpdbe_df[funpdbe_df.chain_id.eq(record['chain_id'])]
| author_insertion_code | author_residue_number | chain_id | chem_comp_id | confidence_classification | confidence_score | entity_id | evidence_codes | label | origin | pdb_id | raw_score | residue_number | site_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | A | MET | NaN | 0.5 | 1 | ['ECO_0000364', 'ECO_0000203'] | backbone | dynamine | 3ii6 | 0.765000 | 1 | 1 | |
| 1 | 2 | A | GLU | NaN | 0.5 | 1 | ['ECO_0000364', 'ECO_0000203'] | backbone | dynamine | 3ii6 | 0.773000 | 2 | 1 | |
| 2 | 3 | A | ARG | NaN | 0.5 | 1 | ['ECO_0000364', 'ECO_0000203'] | backbone | dynamine | 3ii6 | 0.784000 | 3 | 1 | |
| 3 | 4 | A | LYS | NaN | 0.5 | 1 | ['ECO_0000364', 'ECO_0000203'] | backbone | dynamine | 3ii6 | 0.788000 | 4 | 1 | |
| 4 | 5 | A | ILE | NaN | 0.5 | 1 | ['ECO_0000364', 'ECO_0000203'] | backbone | dynamine | 3ii6 | 0.792000 | 5 | 1 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 9282 | 161 | A | ARG | high | NaN | 1 | ['ECO_0000006', 'ECO_0000088'] | Disease | FoldX | 3ii6 | 1.124540 | 161 | 1 | |
| 9283 | 56 | A | ALA | high | NaN | 1 | ['ECO_0000006', 'ECO_0000088'] | Polymorphism | FoldX | 3ii6 | 2.279820 | 56 | 2 | |
| 9284 | 12 | A | SER | high | NaN | 1 | ['ECO_0000006', 'ECO_0000088'] | Polymorphism | FoldX | 3ii6 | 0.314961 | 12 | 3 | |
| 9285 | 43 | A | TRP | high | NaN | 1 | ['ECO_0000006', 'ECO_0000088'] | Disease | FoldX | 3ii6 | 2.777570 | 43 | 4 | |
| 9286 | 142 | A | GLU | high | NaN | 1 | ['ECO_0000006', 'ECO_0000088'] | Polymorphism | FoldX | 3ii6 | -0.196969 | 142 | 5 |
1613 rows × 14 columns
Collecting Chain|Residue-Level Functional Annotation From SIFTS API | PDBe Graph API
Jose M Dana, Aleksandras Gutmanas, Nidhi Tyagi, Guoying Qi, Claire O’Donovan, Maria Martin, Sameer Velankar, SIFTS: updated Structure Integration with Function, Taxonomy and Sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins, Nucleic Acids Research, Volume 47, Issue D1, 08 January 2019, Pages D482–D489, https://doi.org/10.1093/nar/gky1114
Structure Integration with Function, Taxonomy and Sequence (SIFTS) is a project in the PDBe-KB resource for residue-level mapping between UniProt and PDB entries. SIFTS also provides annotation from the IntEnz, GO, InterPro, Pfam, CATH, SCOP, PubMed, Ensembl and Homologene resources. The information is updated and released every week concurrently with the release of new PDB entries and is widely used by resources such as RCSB PDB, PDBj, PDBsum, Pfam, SCOP and InterPro.
 |
api/mappings/orgraph-api/mappings/api/mappings/sequence_domains/- NOTE: (interpro+pfam)
api/mappings/interpro/api/mappings/pfam/
api/mappings/structural_domains/- NOTE: (scop+cath)
api/mappings/scop/api/mappings/cath/
api/mappings/cath_b/api/mappings/go/(chain-level)api/mappings/ec/(chain-level)api/mappings/hmmer/
api/pdb/entry/secondary_structure/graph-api/pdb/sequence_conservation/
pdb_ob.fetch_from_pdbe_api('api/mappings/interpro/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| InterPro | chain_id | end | entity_id | identifier | name | pdb_id | start | struct_asym_id | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | IPR009089 | A | {"author_residue_number":117,"author_insertion... | 1 | XRCC4, N-terminal domain superfamily | XRCC4, N-terminal domain superfamily | 3ii6 | {"author_residue_number":1,"author_insertion_c... | A |
| 22 | IPR010585 | A | {"author_residue_number":200,"author_insertion... | 1 | DNA repair protein XRCC4 | DNA repair protein XRCC4 | 3ii6 | {"author_residue_number":1,"author_insertion_c... | A |
| 23 | IPR010585 | A | {"author_residue_number":201,"author_insertion... | 1 | DNA repair protein XRCC4 | DNA repair protein XRCC4 | 3ii6 | {"author_residue_number":2,"author_insertion_c... | A |
pdb_ob.fetch_from_pdbe_api('api/mappings/pfam/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| Pfam | chain_id | coverage | description | end | entity_id | identifier | name | pdb_id | start | struct_asym_id |
|---|
pdb_ob.fetch_from_pdbe_api('api/mappings/structural_domains/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| CATH | architecture | chain_id | class | domain | end | entity_id | homology | identifier | name | pdb_id | segment_id | start | struct_asym_id | topology | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 1.20.5.370 | Up-down Bundle | A | Mainly Alpha | 3ii6A02 | {"author_residue_number":176,"author_insertion... | 1 | Single alpha-helices involved in coiled-coils ... | Single alpha-helices involved in coiled-coils ... | Dna repair protein xrcc4. Chain: a, b, c, d. F... | 3ii6 | 1 | {"author_residue_number":119,"author_insertion... | A | Single alpha-helices involved in coiled-coils ... |
| 8 | 2.170.210.10 | Beta Complex | A | Mainly Beta | 3ii6A01 | {"author_residue_number":118,"author_insertion... | 1 | DNA double-strand break repair and VJ recombin... | Dna Repair Protein Xrcc4; Chain: A, domain 1 | Dna repair protein xrcc4. Chain: a, b, c, d. F... | 3ii6 | 1 | {"author_residue_number":1,"author_insertion_c... | A | Dna Repair Protein Xrcc4; Chain: A, domain 1 |
pdb_ob.fetch_from_pdbe_api('api/mappings/cath_b/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| CATH-B | architecture | chain_id | class | domain | end | entity_id | homology | identifier | name | pdb_id | segment_id | start | struct_asym_id | topology | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 1.20.5.370 | Up-down Bundle | A | Mainly Alpha | 3ii6A02 | {"author_residue_number":176,"author_insertion... | 1 | Single alpha-helices involved in coiled-coils ... | Single alpha-helices involved in coiled-coils ... | Dna repair protein xrcc4. Chain: a, b, c, d. F... | 3ii6 | 1 | {"author_residue_number":119,"author_insertion... | A | Single alpha-helices involved in coiled-coils ... |
| 8 | 2.170.210.10 | Beta Complex | A | Mainly Beta | 3ii6A01 | {"author_residue_number":118,"author_insertion... | 1 | DNA double-strand break repair and VJ recombin... | Dna Repair Protein Xrcc4; Chain: A, domain 1 | Dna repair protein xrcc4. Chain: a, b, c, d. F... | 3ii6 | 1 | {"author_residue_number":1,"author_insertion_c... | A | Dna Repair Protein Xrcc4; Chain: A, domain 1 |
pdb_ob.fetch_from_pdbe_api('api/mappings/go/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| GO | category | chain_id | definition | entity_id | identifier | name | pdb_id | struct_asym_id | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | GO:0006310 | Biological_process | A | Any process in which a new genotype is formed ... | 1 | DNA recombination | DNA recombination | 3ii6 | A |
| 4 | GO:0006302 | Biological_process | A | The repair of double-strand breaks in DNA via ... | 1 | double-strand break repair | double-strand break repair | 3ii6 | A |
| 10 | GO:0005634 | Cellular_component | A | A membrane-bounded organelle of eukaryotic cel... | 1 | nucleus | nucleus | 3ii6 | A |
| 18 | GO:0003677 | Molecular_function | A | Any molecular function by which a gene product... | 1 | DNA binding | DNA binding | 3ii6 | A |
pdb_ob.fetch_from_pdbe_api('api/mappings/ec/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| EC | accepted_name | chain_id | entity_id | identifier | pdb_id | reaction | struct_asym_id | synonyms | systematic_name |
|---|
pdb_ob.fetch_from_pdbe_api('api/mappings/hmmer/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| HMMER | chain_id | coverage | description | end | entity_id | hmm_end | hmm_length | hmm_start | identifier | name | pdb_id | start | struct_asym_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | PF06632 | A | 0.608 | DNA double-strand break repair and V(D)J recom... | {"author_residue_number":200,"author_insertion... | 1 | 205 | 337 | 1 | DNA double-strand break repair and V(D)J recom... | XRCC4 | 3ii6 | {"author_residue_number":1,"author_insertion_c... | A |
pdb_ob.fetch_from_pdbe_api('api/pdb/entry/secondary_structure/', Base.to_dataframe).result().query('chain_id == "{}"'.format(record['chain_id']))
| chain_id | end | entity_id | pdb_id | secondary_structure | sheet_id | start | struct_asym_id | |
|---|---|---|---|---|---|---|---|---|
| 0 | A | {"author_residue_number":59,"author_insertion_... | 1 | 3ii6 | helices | NaN | {"author_residue_number":49,"author_insertion_... | A |
| 1 | A | {"author_residue_number":75,"author_insertion_... | 1 | 3ii6 | helices | NaN | {"author_residue_number":62,"author_insertion_... | A |
| 2 | A | {"author_residue_number":201,"author_insertion... | 1 | 3ii6 | helices | NaN | {"author_residue_number":118,"author_insertion... | A |
| 3 | A | {"author_residue_number":10,"author_insertion_... | 1 | 3ii6 | strands | 1.0 | {"author_residue_number":3,"author_insertion_c... | A |
| 4 | A | {"author_residue_number":23,"author_insertion_... | 1 | 3ii6 | strands | 1.0 | {"author_residue_number":13,"author_insertion_... | A |
| 5 | A | {"author_residue_number":37,"author_insertion_... | 1 | 3ii6 | strands | 1.0 | {"author_residue_number":31,"author_insertion_... | A |
| 6 | A | {"author_residue_number":44,"author_insertion_... | 1 | 3ii6 | strands | 1.0 | {"author_residue_number":42,"author_insertion_... | A |
| 7 | A | {"author_residue_number":48,"author_insertion_... | 1 | 3ii6 | strands | 1.0 | {"author_residue_number":46,"author_insertion_... | A |
| 8 | A | {"author_residue_number":88,"author_insertion_... | 1 | 3ii6 | strands | 2.0 | {"author_residue_number":84,"author_insertion_... | A |
| 9 | A | {"author_residue_number":100,"author_insertion... | 1 | 3ii6 | strands | 2.0 | {"author_residue_number":94,"author_insertion_... | A |
| 10 | A | {"author_residue_number":112,"author_insertion... | 1 | 3ii6 | strands | 2.0 | {"author_residue_number":105,"author_insertion... | A |
| 11 | A | {"author_residue_number":115,"author_insertion... | 1 | 3ii6 | strands | 1.0 | {"author_residue_number":114,"author_insertion... | A |
seq_conser_df = pdb_ob.fetch_from_pdbe_api(
'graph-api/pdb/sequence_conservation/',
Base.to_dataframe,
mask_id="%s/%s" % (record['pdb_id'], record['entity_id'])
).result()
seq_conser_df
| conservation_score | entity_id | length | letter_array | pdb_id | proba_array | residue_number | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 203 | ["M","L","I","V","A","F","T","S","K","R","E","... | 3ii6 | [0.217,0.168,0.096,0.096,0.054,0.042,0.04,0.03... | 1 |
| 1 | 0 | 1 | 203 | ["E","D","K","S","N","A","Q","R","T","G","L","... | 3ii6 | [0.239,0.101,0.082,0.071,0.065,0.062,0.062,0.0... | 2 |
| 2 | 0 | 1 | 203 | ["R","K","E","T","S","A","Q","N","D","G","L","... | 3ii6 | [0.158,0.151,0.08,0.071,0.07,0.064,0.064,0.049... | 3 |
| 3 | 0 | 1 | 203 | ["K","S","R","A","E","T","Q","N","D","V","L","... | 3ii6 | [0.121,0.097,0.089,0.088,0.087,0.085,0.064,0.0... | 4 |
| 4 | 2 | 1 | 203 | ["V","I","L","A","M","T","F","S","C","Y","E","... | 3ii6 | [0.444,0.227,0.128,0.033,0.029,0.026,0.021,0.0... | 5 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 197 | 0 | 1 | 203 | ["L","V","I","A","T","K","S","Q","E","F","M","... | 3ii6 | [0.286,0.099,0.079,0.07,0.051,0.046,0.046,0.03... | 198 |
| 198 | 0 | 1 | 203 | ["L","A","K","E","S","V","T","R","I","Q","N","... | 3ii6 | [0.156,0.08,0.075,0.071,0.068,0.067,0.066,0.05... | 199 |
| 199 | 0 | 1 | 203 | ["N","S","E","K","A","D","Q","R","T","V","L","... | 3ii6 | [0.109,0.106,0.103,0.094,0.077,0.068,0.067,0.0... | 200 |
| 200 | 0 | 1 | 203 | ["E","A","K","S","D","T","N","Q","V","R","L","... | 3ii6 | [0.142,0.093,0.081,0.074,0.072,0.065,0.057,0.0... | 201 |
| 201 | 0 | 1 | 203 | ["A","V","S","L","I","T","K","E","G","D","R","... | 3ii6 | [0.136,0.094,0.083,0.082,0.071,0.066,0.055,0.0... | 202 |
202 rows × 7 columns
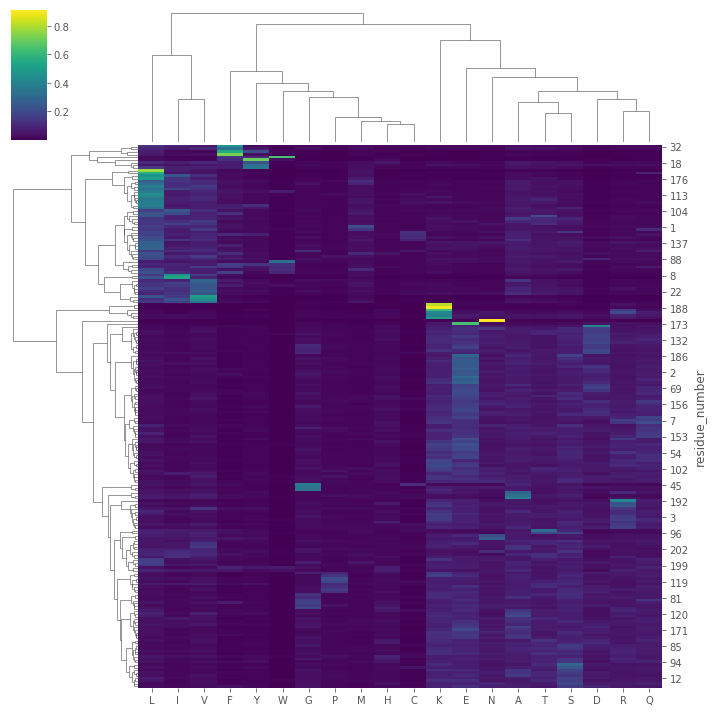
Visualization
import matplotlib.pyplot as plt
import seaborn as sns
import orjson as json
plt.style.use('ggplot')
expanded_seq_conser_df = DataFrame(
seq_conser_df.apply(lambda x: dict(zip(json.loads(x['letter_array']), json.loads(x['proba_array']))), axis=1).tolist(),
index=seq_conser_df.residue_number
)
expanded_seq_conser_df
| M | L | I | V | A | F | T | S | K | R | E | Q | G | Y | N | D | P | H | C | W | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| residue_number | ||||||||||||||||||||
| 1 | 0.217 | 0.168 | 0.096 | 0.096 | 0.054 | 0.042 | 0.040 | 0.039 | 0.036 | 0.029 | 0.028 | 0.024 | 0.023 | 0.023 | 0.021 | 0.019 | 0.014 | 0.013 | 0.011 | 0.007 |
| 2 | 0.013 | 0.032 | 0.021 | 0.030 | 0.062 | 0.012 | 0.047 | 0.071 | 0.082 | 0.051 | 0.239 | 0.062 | 0.041 | 0.015 | 0.065 | 0.101 | 0.023 | 0.025 | 0.006 | 0.004 |
| 3 | 0.017 | 0.038 | 0.024 | 0.034 | 0.064 | 0.013 | 0.071 | 0.070 | 0.151 | 0.158 | 0.080 | 0.064 | 0.045 | 0.016 | 0.049 | 0.045 | 0.021 | 0.028 | 0.007 | 0.004 |
| 4 | 0.019 | 0.042 | 0.031 | 0.044 | 0.088 | 0.015 | 0.085 | 0.097 | 0.121 | 0.089 | 0.087 | 0.064 | 0.032 | 0.018 | 0.053 | 0.050 | 0.021 | 0.032 | 0.007 | 0.005 |
| 5 | 0.029 | 0.128 | 0.227 | 0.444 | 0.033 | 0.021 | 0.026 | 0.013 | 0.008 | 0.007 | 0.008 | 0.007 | 0.007 | 0.009 | 0.006 | 0.005 | 0.006 | 0.004 | 0.010 | 0.003 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 198 | 0.034 | 0.286 | 0.079 | 0.099 | 0.070 | 0.034 | 0.051 | 0.046 | 0.046 | 0.030 | 0.038 | 0.039 | 0.022 | 0.026 | 0.025 | 0.019 | 0.018 | 0.017 | 0.014 | 0.007 |
| 199 | 0.028 | 0.156 | 0.051 | 0.067 | 0.080 | 0.027 | 0.066 | 0.068 | 0.075 | 0.059 | 0.071 | 0.051 | 0.029 | 0.023 | 0.044 | 0.036 | 0.023 | 0.023 | 0.015 | 0.007 |
| 200 | 0.018 | 0.042 | 0.027 | 0.043 | 0.077 | 0.016 | 0.056 | 0.106 | 0.094 | 0.058 | 0.103 | 0.067 | 0.036 | 0.017 | 0.109 | 0.068 | 0.023 | 0.029 | 0.008 | 0.005 |
| 201 | 0.020 | 0.048 | 0.037 | 0.056 | 0.093 | 0.018 | 0.065 | 0.074 | 0.081 | 0.055 | 0.142 | 0.056 | 0.045 | 0.018 | 0.057 | 0.072 | 0.024 | 0.025 | 0.010 | 0.005 |
| 202 | 0.025 | 0.082 | 0.071 | 0.094 | 0.136 | 0.029 | 0.066 | 0.083 | 0.055 | 0.043 | 0.053 | 0.036 | 0.052 | 0.022 | 0.041 | 0.043 | 0.026 | 0.019 | 0.017 | 0.006 |
202 rows × 20 columns
plt.figure(figsize=(10,8))
sns.heatmap(expanded_seq_conser_df, cmap='viridis')

sns.clustermap(expanded_seq_conser_df, cmap='viridis', method='ward')